Resolver Architecture Concept#
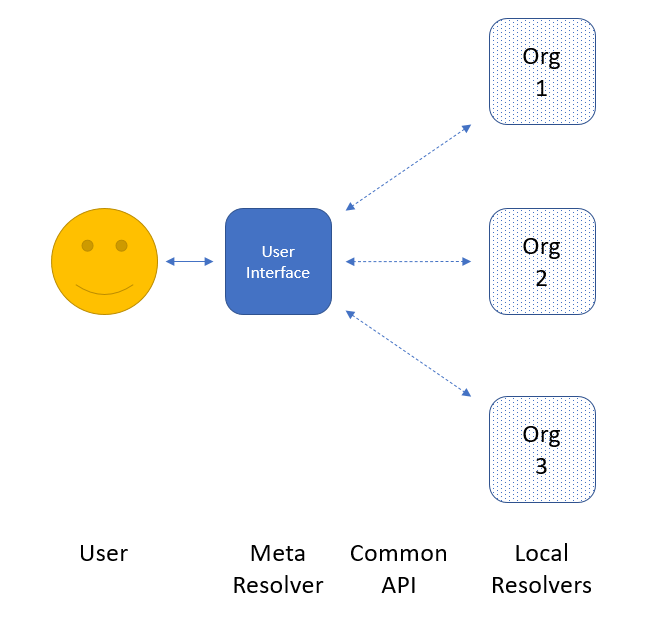
The proposed design of the global resolver is fairly straightforward, and has three main components: a shared data model for information exchange, a local resolver implemented and hosted by each participating organization, and a central “meta resolver” that communicates with the local resolvers and provides the web page interface that the user interacts with directly. In brief, the user would enter a query, and the meta resolver would send that query to each local resolver in its list (presumably in parallel). Each local resolver would perform the lookup within their system, and respond back to the meta resolver which would gather all the responses to present to the user. The most important part would be for the user to be able to clearly understand which organizations are responding, and to have links to follow through to those resources for more detail. In this way, the meta resolver does not need to know what each resource’s full record contains, or to try to present each such heterogeneous record in a common way. All it needs to know is “this organization has information on this chemical” and it is then up to the user to decide where to go from there.
Common Data Model#
If this system is to be practical to implement across multiple databases all over the world, with new participants to be added at any time and without great effort, there must be a common “language” that is used for communications between the central meta resolver and the individual organizations’ local resolvers. It should use generic common standards for communications and the data model, that are not dependent on any particular host platform or programming language or database system. This simplifies the meta resolver because it uses exactly the same method to communicate with each organization, and frees individual organizations to implement their local component in whatever way is most convenient for them given their existing infrastructure.
What is proposed here – but is certainly open to change – uses the HTTP protocol for communications, with normal CGI arguments for input, and then a simple JSON construct for the response data. These two standards are universal enough that programmers using just about any platform should have access to the data parsers and communications tools needed to implement this service.
Parenthetical note: PubChem has a very simple data model for this written in XML schema, as NCBI has (publicly available) software to read and write equivalent JSON or XML (or ASN.1 historically used by NCBI) based on a single schema. This schema, in its prototype form, is used to construct the examples in the next section, and is available at
https://pubchem.ncbi.nlm.nih.gov/resolver/resolver_data.xsd
But the final implementation, if it uses JSON, should probably use a JSON-native format like JSON Schema (https://json-schema.org/draft/2020-12/json-schema-core.html - although that is still under development). Or, the data format of choice for this whole system could be XML instead. Regardless, there should be some way to make it clear to developers what the structure of the data should be, and make it possible to validate the JSON data automatically. These details, and whether to use HTTP or some other protocol, are not inherently critical to the overall design of this system, but would be important to work out before individual organizations begin work on their implementations.
