CAS Common Chemistry API#
About this interactive  recipe
recipe
Author: Vincent Scalfani
Reviewer: Stuart Chalk
Topics: How to interact with the CAS Common Chemistry API using Python.
Attribution: Adapted from the MIT licensed University of Alabama Scholarly API Cookbook Use of the CAS Common Chemistry API requires registration: https://www.cas.org/services/commonchemistry-api. Example data shown is credited to CAS Common Chemistry, which is licensed under the CC BY-NC 4.0 license.
Format: Interactive Jupyter Notebook (Python)
Scenarios: You are search for identifiers and general property of common chemical substances.
Skills: You should be familiar with
Learning outcomes: After completing this example you should understand:
What kind of data is available through the CAS Common Chemistry API
How to interact with the CAS Common Chemistry API using Python
Citation: ‘CAS Common Chemistry API’, Vincent Scalfani, The IUPAC FAIR Chemistry Cookbook, Contributed: 2024-02-14 https://w3id.org/ifcc/IFCC011.
Reuse: This notebook is made available under a CC-BY-4.0 license.
1. Common Chemistry Record Detail Retrieval#
Information about substances in CAS Common Chemistry can be retrieved using the /detail API and a CAS RN identifier:
Import libraries#
import requests
from pprint import pprint
Setup API parameters#
detail_base_url = "https://commonchemistry.cas.org/api/detail?"
casrn1 = "10094-36-7" # ethyl cyclohexanepropionate
Request data from CAS Common Chemistry Detail API#
casrn1_data = requests.get(detail_base_url + "cas_rn=" + casrn1).json()
pprint(casrn1_data)
{'message': 'API key required. Please include a valid API key in the '
"'X-API-KEY' header."}
Display the Molecule Drawing#
# get svg image text
svg_string1 = casrn1_data["image"]
# display the molecule
from IPython.display import SVG
SVG(svg_string1)
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
Cell In[4], line 2
1 # get svg image text
----> 2 svg_string1 = casrn1_data["image"]
4 # display the molecule
5 from IPython.display import SVG
KeyError: 'image'
Select some specific data#
# Get Experimental Properties
casrn1_data["experimentalProperties"][0]
{'name': 'Boiling Point',
'property': '105-113 °C @ Press: 17 Torr',
'sourceNumber': 1}
# Get Boiling Point property
casrn1_data["experimentalProperties"][0]["property"]
'105-113 °C @ Press: 17 Torr'
# Get InChIKey
casrn1_data["inchiKey"]
'InChIKey=NRVPMFHPHGBQLP-UHFFFAOYSA-N'
# Get Canonical SMILES
casrn1_data["canonicalSmile"]
'O=C(OCC)CCC1CCCCC1'
2. Common Chemistry API record detail retrieval in a loop#
Import libraries#
import requests
from pprint import pprint
from time import sleep
Setup API parameters#
detail_base_url = "https://commonchemistry.cas.org/api/detail?"
casrn_list = ["10094-36-7", "10031-92-2", "10199-61-8", "10036-21-2", "1019020-13-3"]
Request data for each CAS RN and save to a list#
casrn_data = []
for casrn in casrn_list:
casrn_data.append(requests.get(detail_base_url + "cas_rn=" + casrn).json())
sleep(1) # add a delay between API calls
casrn_data[0:2] # view first 2
[{'uri': 'substance/pt/10094367',
'rn': '10094-36-7',
'name': 'Ethyl cyclohexanepropionate',
'image': '<svg width="228.6" viewBox="0 0 7620 3716" text-rendering="auto" stroke-width="1" stroke-opacity="1" stroke-miterlimit="10" stroke-linejoin="miter" stroke-linecap="square" stroke-dashoffset="0" stroke-dasharray="none" stroke="black" shape-rendering="auto" image-rendering="auto" height="111.48" font-weight="normal" font-style="normal" font-size="12" font-family="\'Dialog\'" fill-opacity="1" fill="black" color-rendering="auto" color-interpolation="auto" xmlns="http://www.w3.org/2000/svg"><g><g stroke="white" fill="white"><rect y="0" x="0" width="7620" stroke="none" height="3716"/></g><g transform="translate(32866,32758)" text-rendering="geometricPrecision" stroke-width="44" stroke-linejoin="round" stroke-linecap="round"><line y2="-30850" y1="-31419" x2="-30792" x1="-31777" fill="none"/><line y2="-29715" y1="-30850" x2="-30792" x1="-30792" fill="none"/><line y2="-31419" y1="-30850" x2="-31777" x1="-32762" fill="none"/><line y2="-29146" y1="-29715" x2="-31777" x1="-30792" fill="none"/><line y2="-30850" y1="-29715" x2="-32762" x1="-32762" fill="none"/><line y2="-29715" y1="-29146" x2="-32762" x1="-31777" fill="none"/><line y2="-31376" y1="-30850" x2="-29885" x1="-30792" fill="none"/><line y2="-30850" y1="-31376" x2="-28978" x1="-29885" fill="none"/><line y2="-31376" y1="-30850" x2="-28071" x1="-28978" fill="none"/><line y2="-30960" y1="-31376" x2="-27352" x1="-28071" fill="none"/><line y2="-31376" y1="-30960" x2="-26257" x1="-26976" fill="none"/><line y2="-30850" y1="-31376" x2="-25350" x1="-26257" fill="none"/><line y2="-32202" y1="-31376" x2="-28140" x1="-28140" fill="none"/><line y2="-32202" y1="-31376" x2="-28002" x1="-28002" fill="none"/><text y="-30671" xml:space="preserve" x="-27317" stroke="none" font-size="433.3333" font-family="sans-serif">O</text><text y="-32242" xml:space="preserve" x="-28224" stroke="none" font-size="433.3333" font-family="sans-serif">O</text></g></g></svg>',
'inchi': 'InChI=1S/C11H20O2/c1-2-13-11(12)9-8-10-6-4-3-5-7-10/h10H,2-9H2,1H3',
'inchiKey': 'InChIKey=NRVPMFHPHGBQLP-UHFFFAOYSA-N',
'smile': 'C(CC(OCC)=O)C1CCCCC1',
'canonicalSmile': 'O=C(OCC)CCC1CCCCC1',
'molecularFormula': 'C<sub>11</sub>H<sub>20</sub>O<sub>2</sub>',
'molecularMass': '184.28',
'experimentalProperties': [{'name': 'Boiling Point',
'property': '105-113 °C @ Press: 17 Torr',
'sourceNumber': 1}],
'propertyCitations': [{'docUri': 'document/pt/document/22252593',
'sourceNumber': 1,
'source': 'De Benneville, Peter L.; Journal of the American Chemical Society, (1940), 62, 283-7, CAplus'}],
'synonyms': ['Cyclohexanepropanoic acid, ethyl ester',
'Cyclohexanepropionic acid, ethyl ester',
'Ethyl cyclohexanepropionate',
'Ethyl cyclohexylpropanoate',
'Ethyl 3-cyclohexylpropionate',
'Ethyl 3-cyclohexylpropanoate',
'3-Cyclohexylpropionic acid ethyl ester',
'NSC 71463',
'Ethyl 3-cyclohexanepropionate'],
'replacedRns': [],
'hasMolfile': True},
{'uri': 'substance/pt/10031922',
'rn': '10031-92-2',
'name': 'Ethyl 2-nonynoate',
'image': '<svg width="318.24" viewBox="0 0 10608 2283" text-rendering="auto" stroke-width="1" stroke-opacity="1" stroke-miterlimit="10" stroke-linejoin="miter" stroke-linecap="square" stroke-dashoffset="0" stroke-dasharray="none" stroke="black" shape-rendering="auto" image-rendering="auto" height="68.49" font-weight="normal" font-style="normal" font-size="12" font-family="\'Dialog\'" fill-opacity="1" fill="black" color-rendering="auto" color-interpolation="auto" xmlns="http://www.w3.org/2000/svg"><g><g stroke="white" fill="white"><rect y="0" x="0" width="10608" stroke="none" height="2283"/></g><g transform="translate(32866,32758)" text-rendering="geometricPrecision" stroke-width="44" stroke-linejoin="round" stroke-linecap="round"><line y2="-31899" y1="-31899" x2="-26132" x1="-27178" fill="none"/><line y2="-31988" y1="-31988" x2="-26132" x1="-27178" fill="none"/><line y2="-31809" y1="-31809" x2="-26132" x1="-27178" fill="none"/><line y2="-31899" y1="-31899" x2="-28227" x1="-27178" fill="none"/><line y2="-31376" y1="-31899" x2="-29134" x1="-28227" fill="none"/><line y2="-31899" y1="-31376" x2="-30041" x1="-29134" fill="none"/><line y2="-31376" y1="-31899" x2="-30948" x1="-30041" fill="none"/><line y2="-31899" y1="-31376" x2="-31855" x1="-30948" fill="none"/><line y2="-31376" y1="-31899" x2="-32762" x1="-31855" fill="none"/><line y2="-31899" y1="-31899" x2="-25084" x1="-26132" fill="none"/><line y2="-32315" y1="-31899" x2="-24364" x1="-25084" fill="none"/><line y2="-31899" y1="-32315" x2="-23270" x1="-23989" fill="none"/><line y2="-32422" y1="-31899" x2="-22362" x1="-23270" fill="none"/><line y2="-31070" y1="-31899" x2="-25014" x1="-25014" fill="none"/><line y2="-31070" y1="-31899" x2="-25153" x1="-25153" fill="none"/><text y="-32242" xml:space="preserve" x="-24330" stroke="none" font-size="433.3333" font-family="sans-serif">O</text><text y="-30671" xml:space="preserve" x="-25237" stroke="none" font-size="433.3333" font-family="sans-serif">O</text></g></g></svg>',
'inchi': 'InChI=1S/C11H18O2/c1-3-5-6-7-8-9-10-11(12)13-4-2/h3-8H2,1-2H3',
'inchiKey': 'InChIKey=BFZNMUGAZYAMTG-UHFFFAOYSA-N',
'smile': 'C(C#CCCCCCC)(OCC)=O',
'canonicalSmile': 'O=C(C#CCCCCCC)OCC',
'molecularFormula': 'C<sub>11</sub>H<sub>18</sub>O<sub>2</sub>',
'molecularMass': '182.26',
'experimentalProperties': [],
'propertyCitations': [],
'synonyms': ['2-Nonynoic acid, ethyl ester',
'Ethyl 2-nonynoate',
'NSC 190985'],
'replacedRns': [],
'hasMolfile': True}]
Display Molecule Drawings#
from IPython.display import SVG
# get svg image text
svg_strings = []
for svg_idx in range(len(casrn_data)):
svg_strings.append(casrn_data[svg_idx]["image"])
# display the molecules
for svg_string in svg_strings:
display(SVG(svg_string))
Select some specific data#
# Get canonical SMILES
cansmiles = []
for cansmi in range(len(casrn_data)):
cansmiles.append(casrn_data[cansmi]["canonicalSmile"])
print(cansmiles)
['O=C(OCC)CCC1CCCCC1', 'O=C(C#CCCCCCC)OCC', 'O=C(OCC)CN1N=CC=C1', 'O=C(OCC)C1=CC=CC(=C1)CCC(=O)OCC', 'N=C(OCC)C1=CCCCC1']
# Get synonyms
synonyms_list = []
for syn in range(len(casrn_data)):
synonyms_list.append(casrn_data[syn]["synonyms"])
pprint(synonyms_list)
[['Cyclohexanepropanoic acid, ethyl ester',
'Cyclohexanepropionic acid, ethyl ester',
'Ethyl cyclohexanepropionate',
'Ethyl cyclohexylpropanoate',
'Ethyl 3-cyclohexylpropionate',
'Ethyl 3-cyclohexylpropanoate',
'3-Cyclohexylpropionic acid ethyl ester',
'NSC 71463',
'Ethyl 3-cyclohexanepropionate'],
['2-Nonynoic acid, ethyl ester', 'Ethyl 2-nonynoate', 'NSC 190985'],
['1<em>H</em>-Pyrazole-1-acetic acid, ethyl ester',
'Pyrazole-1-acetic acid, ethyl ester',
'Ethyl 1<em>H</em>-pyrazole-1-acetate',
'Ethyl 1-pyrazoleacetate',
'Ethyl 2-(1<em>H</em>-pyrazol-1-yl)acetate'],
['Benzenepropanoic acid, 3-(ethoxycarbonyl)-, ethyl ester',
'Hydrocinnamic acid, <em>m</em>-carboxy-, diethyl ester',
'Ethyl 3-(ethoxycarbonyl)benzenepropanoate'],
['1-Cyclohexene-1-carboximidic acid, ethyl ester',
'Ethyl 1-cyclohexene-1-carboximidate']]
# Transform synonym "list of lists" to a flat list
synonyms_flat = []
for sublist in synonyms_list:
for synonym in sublist:
synonyms_flat.append(synonym)
pprint(synonyms_flat)
['Cyclohexanepropanoic acid, ethyl ester',
'Cyclohexanepropionic acid, ethyl ester',
'Ethyl cyclohexanepropionate',
'Ethyl cyclohexylpropanoate',
'Ethyl 3-cyclohexylpropionate',
'Ethyl 3-cyclohexylpropanoate',
'3-Cyclohexylpropionic acid ethyl ester',
'NSC 71463',
'Ethyl 3-cyclohexanepropionate',
'2-Nonynoic acid, ethyl ester',
'Ethyl 2-nonynoate',
'NSC 190985',
'1<em>H</em>-Pyrazole-1-acetic acid, ethyl ester',
'Pyrazole-1-acetic acid, ethyl ester',
'Ethyl 1<em>H</em>-pyrazole-1-acetate',
'Ethyl 1-pyrazoleacetate',
'Ethyl 2-(1<em>H</em>-pyrazol-1-yl)acetate',
'Benzenepropanoic acid, 3-(ethoxycarbonyl)-, ethyl ester',
'Hydrocinnamic acid, <em>m</em>-carboxy-, diethyl ester',
'Ethyl 3-(ethoxycarbonyl)benzenepropanoate',
'1-Cyclohexene-1-carboximidic acid, ethyl ester',
'Ethyl 1-cyclohexene-1-carboximidate']
3. Common Chemistry Search#
In addition to the /detail API, the CAS Common Chemistry API has a /search method that allows searching by CAS RN, SMILES, InChI/InChIKey, and name.
Import libraries#
import requests
from pprint import pprint
from time import sleep
Setup API Parameters#
search_base_url = "https://commonchemistry.cas.org/api/search?q="
Request data from CAS Common Chemistry Search API#
# keyword search query
quinine_search_data = requests.get(search_base_url + "quinine").json()
pprint(quinine_search_data)
{'count': 1,
'results': [{'image': '<svg width="309.3" viewBox="0 0 10310 5592" '
'text-rendering="auto" stroke-width="1" '
'stroke-opacity="1" stroke-miterlimit="10" '
'stroke-linejoin="miter" stroke-linecap="square" '
'stroke-dashoffset="0" stroke-dasharray="none" '
'stroke="black" shape-rendering="auto" '
'image-rendering="auto" height="167.76" '
'font-weight="normal" font-style="normal" '
'font-size="12" font-family="\'Dialog\'" '
'fill-opacity="1" fill="black" color-rendering="auto" '
'color-interpolation="auto" '
'xmlns="http://www.w3.org/2000/svg"><g><g '
'stroke="white" fill="white"><rect y="0" x="0" '
'width="10310" stroke="none" height="5592"/></g><g '
'transform="translate(32866,32758)" '
'text-rendering="geometricPrecision" stroke-width="44" '
'stroke-linejoin="round" stroke-linecap="round"><line '
'y2="-28559" y1="-28036" x2="-26635" x1="-25742" '
'fill="none"/><line y2="-29819" y1="-28559" x2="-26635" '
'x1="-26635" fill="none"/><line y2="-28036" y1="-28559" '
'x2="-25367" x1="-24474" fill="none"/><line y2="-30451" '
'y1="-29819" x2="-25555" x1="-26635" fill="none"/><line '
'y2="-28559" y1="-29819" x2="-24474" x1="-24474" '
'fill="none"/><line y2="-29504" y1="-28828" x2="-25194" '
'x1="-26005" fill="none"/><line y2="-29819" y1="-30451" '
'x2="-24474" x1="-25555" fill="none"/><line y2="-29082" '
'y1="-28559" x2="-27542" x1="-26635" fill="none"/><line '
'y2="-29819" y1="-30344" x2="-22660" x1="-23567" '
'fill="none"/><line y2="-29700" y1="-30223" x2="-22729" '
'x1="-23636" fill="none"/><line y2="-28779" y1="-29082" '
'x2="-28071" x1="-27542" fill="none"/><line y2="-30703" '
'y1="-30131" x2="-28524" x1="-27542" fill="none"/><line '
'y2="-31850" y1="-30703" x2="-28524" x1="-28524" '
'fill="none"/><line y2="-31705" y1="-30847" x2="-28354" '
'x1="-28354" fill="none"/><line y2="-30131" y1="-30703" '
'x2="-29507" x1="-28524" fill="none"/><line y2="-30131" '
'y1="-30703" x2="-27542" x1="-26560" fill="none"/><line '
'y2="-30347" y1="-30778" x2="-27505" x1="-26768" '
'fill="none"/><line y2="-31850" y1="-32422" x2="-28524" '
'x1="-29507" fill="none"/><line y2="-32312" y1="-31850" '
'x2="-27730" x1="-28524" fill="none"/><line y2="-30703" '
'y1="-30131" x2="-30489" x1="-29507" fill="none"/><line '
'y2="-30778" y1="-30347" x2="-30281" x1="-29544" '
'fill="none"/><line y2="-30703" y1="-31850" x2="-26560" '
'x1="-26560" fill="none"/><line y2="-32422" y1="-31850" '
'x2="-29507" x1="-30489" fill="none"/><line y2="-32205" '
'y1="-31774" x2="-29544" x1="-30281" fill="none"/><line '
'y2="-31850" y1="-32312" x2="-26560" x1="-27354" '
'fill="none"/><line y2="-31760" y1="-32107" x2="-26745" '
'x1="-27340" fill="none"/><line y2="-31850" y1="-30703" '
'x2="-30489" x1="-30489" fill="none"/><line y2="-30275" '
'y1="-30703" x2="-31200" x1="-30489" fill="none"/><line '
'y2="-30541" y1="-30272" x2="-32040" x1="-31575" '
'fill="none"/><polygon stroke-width="1" stroke="none" '
'points=" -24474 -29819 -23602 -30402 -23532 '
'-30284"/><polygon stroke-width="1" points=" -24474 '
'-29819 -23602 -30402 -23532 -30284" '
'fill="none"/><polygon stroke-width="1" stroke="none" '
'points=" -26635 -28559 -26973 -27837 -27092 '
'-27903"/><polygon stroke-width="1" points=" -26635 '
'-28559 -26973 -27837 -27092 -27903" fill="none"/><line '
'y2="-28860" y1="-28796" x2="-25945" x1="-26066" '
'fill="none"/><line y2="-28657" y1="-28611" x2="-25865" '
'x1="-25952" fill="none"/><line y2="-28454" y1="-28427" '
'x2="-25785" x1="-25838" fill="none"/><line y2="-28252" '
'y1="-28242" x2="-25706" x1="-25723" fill="none"/><line '
'y2="-29478" y1="-29530" x2="-25257" x1="-25130" '
'fill="none"/><line y2="-29686" y1="-29727" x2="-25321" '
'x1="-25221" fill="none"/><line y2="-29894" y1="-29924" '
'x2="-25384" x1="-25312" fill="none"/><line y2="-30102" '
'y1="-30121" x2="-25448" x1="-25403" fill="none"/><line '
'y2="-30310" y1="-30317" x2="-25512" x1="-25493" '
'fill="none"/><line y2="-30131" y1="-30128" x2="-27473" '
'x1="-27612" fill="none"/><line y2="-29914" y1="-29912" '
'x2="-27487" x1="-27598" fill="none"/><line y2="-29697" '
'y1="-29695" x2="-27502" x1="-27583" fill="none"/><line '
'y2="-29480" y1="-29479" x2="-27516" x1="-27569" '
'fill="none"/><line y2="-29263" y1="-29263" x2="-27530" '
'x1="-27554" fill="none"/><text y="-28380" '
'xml:space="preserve" x="-28602" stroke="none" '
'font-size="433.3333" '
'font-family="sans-serif">OH</text><text y="-29983" '
'xml:space="preserve" x="-31540" stroke="none" '
'font-size="433.3333" '
'font-family="sans-serif">O</text><text y="-30691" '
'xml:space="preserve" x="-32762" stroke="none" '
'font-size="433.3333" '
'font-family="sans-serif">CH</text><text y="-30602" '
'xml:space="preserve" x="-32185" stroke="none" '
'font-size="313.3333" '
'font-family="sans-serif">3</text><text y="-32242" '
'xml:space="preserve" x="-27695" stroke="none" '
'font-size="433.3333" '
'font-family="sans-serif">N</text><text y="-27747" '
'xml:space="preserve" x="-25708" stroke="none" '
'font-size="433.3333" '
'font-family="sans-serif">N</text><text y="-27473" '
'xml:space="preserve" x="-27311" stroke="none" '
'font-size="433.3333" '
'font-family="sans-serif">H</text><text y="-28600" '
'xml:space="preserve" x="-27695" stroke="none" '
'font-style="italic" font-size="313.3333" '
'font-family="sans-serif">R</text><text y="-28522" '
'xml:space="preserve" x="-26540" stroke="none" '
'font-style="italic" font-size="313.3333" '
'font-family="sans-serif">S</text><text y="-27337" '
'xml:space="preserve" x="-25818" stroke="none" '
'font-style="italic" font-size="313.3333" '
'font-family="sans-serif">S</text><text y="-30573" '
'xml:space="preserve" x="-25708" stroke="none" '
'font-style="italic" font-size="313.3333" '
'font-family="sans-serif">S</text><text y="-29495" '
'xml:space="preserve" x="-24876" stroke="none" '
'font-style="italic" font-size="313.3333" '
'font-family="sans-serif">R</text></g></g></svg>',
'name': 'Quinine',
'rn': '130-95-0'}]}
Note that with the CAS Common Chemistry Search API, only the image data, name, and CAS RN is returned. In order to retrieve the full record, we can combine our search with the related detail API:
# search query
quinine_search_data = requests.get(search_base_url + "quinine").json()
# extract our CAS RN
quinine_rn = quinine_search_data["results"][0]["rn"]
print(quinine_rn)
130-95-0
# get detailed record for quinine
detail_base_url = "https://commonchemistry.cas.org/api/detail?"
quinine_detail_data = requests.get(detail_base_url + "cas_rn=" + quinine_rn).json()
pprint(quinine_detail_data)
{'canonicalSmile': 'OC(C=1C=CN=C2C=CC(OC)=CC21)C3N4CCC(C3)C(C=C)C4',
'experimentalProperties': [{'name': 'Melting Point',
'property': '177 °C (decomp)',
'sourceNumber': 1}],
'hasMolfile': True,
'image': '<svg width="309.3" viewBox="0 0 10310 5592" text-rendering="auto" '
'stroke-width="1" stroke-opacity="1" stroke-miterlimit="10" '
'stroke-linejoin="miter" stroke-linecap="square" '
'stroke-dashoffset="0" stroke-dasharray="none" stroke="black" '
'shape-rendering="auto" image-rendering="auto" height="167.76" '
'font-weight="normal" font-style="normal" font-size="12" '
'font-family="\'Dialog\'" fill-opacity="1" fill="black" '
'color-rendering="auto" color-interpolation="auto" '
'xmlns="http://www.w3.org/2000/svg"><g><g stroke="white" '
'fill="white"><rect y="0" x="0" width="10310" stroke="none" '
'height="5592"/></g><g transform="translate(32866,32758)" '
'text-rendering="geometricPrecision" stroke-width="44" '
'stroke-linejoin="round" stroke-linecap="round"><line y2="-28559" '
'y1="-28036" x2="-26635" x1="-25742" fill="none"/><line y2="-29819" '
'y1="-28559" x2="-26635" x1="-26635" fill="none"/><line y2="-28036" '
'y1="-28559" x2="-25367" x1="-24474" fill="none"/><line y2="-30451" '
'y1="-29819" x2="-25555" x1="-26635" fill="none"/><line y2="-28559" '
'y1="-29819" x2="-24474" x1="-24474" fill="none"/><line y2="-29504" '
'y1="-28828" x2="-25194" x1="-26005" fill="none"/><line y2="-29819" '
'y1="-30451" x2="-24474" x1="-25555" fill="none"/><line y2="-29082" '
'y1="-28559" x2="-27542" x1="-26635" fill="none"/><line y2="-29819" '
'y1="-30344" x2="-22660" x1="-23567" fill="none"/><line y2="-29700" '
'y1="-30223" x2="-22729" x1="-23636" fill="none"/><line y2="-28779" '
'y1="-29082" x2="-28071" x1="-27542" fill="none"/><line y2="-30703" '
'y1="-30131" x2="-28524" x1="-27542" fill="none"/><line y2="-31850" '
'y1="-30703" x2="-28524" x1="-28524" fill="none"/><line y2="-31705" '
'y1="-30847" x2="-28354" x1="-28354" fill="none"/><line y2="-30131" '
'y1="-30703" x2="-29507" x1="-28524" fill="none"/><line y2="-30131" '
'y1="-30703" x2="-27542" x1="-26560" fill="none"/><line y2="-30347" '
'y1="-30778" x2="-27505" x1="-26768" fill="none"/><line y2="-31850" '
'y1="-32422" x2="-28524" x1="-29507" fill="none"/><line y2="-32312" '
'y1="-31850" x2="-27730" x1="-28524" fill="none"/><line y2="-30703" '
'y1="-30131" x2="-30489" x1="-29507" fill="none"/><line y2="-30778" '
'y1="-30347" x2="-30281" x1="-29544" fill="none"/><line y2="-30703" '
'y1="-31850" x2="-26560" x1="-26560" fill="none"/><line y2="-32422" '
'y1="-31850" x2="-29507" x1="-30489" fill="none"/><line y2="-32205" '
'y1="-31774" x2="-29544" x1="-30281" fill="none"/><line y2="-31850" '
'y1="-32312" x2="-26560" x1="-27354" fill="none"/><line y2="-31760" '
'y1="-32107" x2="-26745" x1="-27340" fill="none"/><line y2="-31850" '
'y1="-30703" x2="-30489" x1="-30489" fill="none"/><line y2="-30275" '
'y1="-30703" x2="-31200" x1="-30489" fill="none"/><line y2="-30541" '
'y1="-30272" x2="-32040" x1="-31575" fill="none"/><polygon '
'stroke-width="1" stroke="none" points=" -24474 -29819 -23602 -30402 '
'-23532 -30284"/><polygon stroke-width="1" points=" -24474 -29819 '
'-23602 -30402 -23532 -30284" fill="none"/><polygon stroke-width="1" '
'stroke="none" points=" -26635 -28559 -26973 -27837 -27092 '
'-27903"/><polygon stroke-width="1" points=" -26635 -28559 -26973 '
'-27837 -27092 -27903" fill="none"/><line y2="-28860" y1="-28796" '
'x2="-25945" x1="-26066" fill="none"/><line y2="-28657" y1="-28611" '
'x2="-25865" x1="-25952" fill="none"/><line y2="-28454" y1="-28427" '
'x2="-25785" x1="-25838" fill="none"/><line y2="-28252" y1="-28242" '
'x2="-25706" x1="-25723" fill="none"/><line y2="-29478" y1="-29530" '
'x2="-25257" x1="-25130" fill="none"/><line y2="-29686" y1="-29727" '
'x2="-25321" x1="-25221" fill="none"/><line y2="-29894" y1="-29924" '
'x2="-25384" x1="-25312" fill="none"/><line y2="-30102" y1="-30121" '
'x2="-25448" x1="-25403" fill="none"/><line y2="-30310" y1="-30317" '
'x2="-25512" x1="-25493" fill="none"/><line y2="-30131" y1="-30128" '
'x2="-27473" x1="-27612" fill="none"/><line y2="-29914" y1="-29912" '
'x2="-27487" x1="-27598" fill="none"/><line y2="-29697" y1="-29695" '
'x2="-27502" x1="-27583" fill="none"/><line y2="-29480" y1="-29479" '
'x2="-27516" x1="-27569" fill="none"/><line y2="-29263" y1="-29263" '
'x2="-27530" x1="-27554" fill="none"/><text y="-28380" '
'xml:space="preserve" x="-28602" stroke="none" font-size="433.3333" '
'font-family="sans-serif">OH</text><text y="-29983" '
'xml:space="preserve" x="-31540" stroke="none" font-size="433.3333" '
'font-family="sans-serif">O</text><text y="-30691" '
'xml:space="preserve" x="-32762" stroke="none" font-size="433.3333" '
'font-family="sans-serif">CH</text><text y="-30602" '
'xml:space="preserve" x="-32185" stroke="none" font-size="313.3333" '
'font-family="sans-serif">3</text><text y="-32242" '
'xml:space="preserve" x="-27695" stroke="none" font-size="433.3333" '
'font-family="sans-serif">N</text><text y="-27747" '
'xml:space="preserve" x="-25708" stroke="none" font-size="433.3333" '
'font-family="sans-serif">N</text><text y="-27473" '
'xml:space="preserve" x="-27311" stroke="none" font-size="433.3333" '
'font-family="sans-serif">H</text><text y="-28600" '
'xml:space="preserve" x="-27695" stroke="none" font-style="italic" '
'font-size="313.3333" font-family="sans-serif">R</text><text '
'y="-28522" xml:space="preserve" x="-26540" stroke="none" '
'font-style="italic" font-size="313.3333" '
'font-family="sans-serif">S</text><text y="-27337" '
'xml:space="preserve" x="-25818" stroke="none" font-style="italic" '
'font-size="313.3333" font-family="sans-serif">S</text><text '
'y="-30573" xml:space="preserve" x="-25708" stroke="none" '
'font-style="italic" font-size="313.3333" '
'font-family="sans-serif">S</text><text y="-29495" '
'xml:space="preserve" x="-24876" stroke="none" font-style="italic" '
'font-size="313.3333" '
'font-family="sans-serif">R</text></g></g></svg>',
'inchi': 'InChI=1S/C20H24N2O2/c1-3-13-12-22-9-7-14(13)10-19(22)20(23)16-6-8-21-18-5-4-15(24-2)11-17(16)18/h3-6,8,11,13-14,19-20,23H,1,7,9-10,12H2,2H3/t13-,14-,19-,20+/m0/s1',
'inchiKey': 'InChIKey=LOUPRKONTZGTKE-WZBLMQSHSA-N',
'molecularFormula': 'C<sub>20</sub>H<sub>24</sub>N<sub>2</sub>O<sub>2</sub>',
'molecularMass': '324.42',
'name': 'Quinine',
'propertyCitations': [{'docUri': '',
'source': 'Drugs - Synonyms and Properties data were '
'obtained from Ashgate Publishing Co. (US)',
'sourceNumber': 1}],
'replacedRns': ['6912-57-8',
'12239-42-8',
'21480-31-9',
'55980-20-6',
'72646-90-3',
'95650-40-1',
'128544-03-6',
'767303-40-2',
'840482-04-4',
'857212-53-4',
'864908-93-0',
'875538-34-4',
'888714-03-2',
'890027-24-4',
'894767-09-0',
'898813-59-7',
'898814-28-3',
'899813-83-3',
'900786-66-5',
'900789-95-9',
'906550-97-8',
'909263-47-4',
'909767-48-2',
'909882-78-6',
'910878-25-0',
'910880-97-6',
'911445-75-5',
'918778-04-8',
'1071756-51-8',
'1267651-57-9',
'1628705-47-4',
'2244812-93-7',
'2244812-97-1',
'2409557-51-1',
'2566761-34-8'],
'rn': '130-95-0',
'smile': '[C@@H](O)(C=1C2=C(C=CC(OC)=C2)N=CC1)[C@]3([N@@]4C[C@H](C=C)[C@](C3)(CC4)[H])[H]',
'synonyms': ['Cinchonan-9-ol, 6′-methoxy-, (8α,9<em>R</em>)-',
'Quinine',
'(8α,9<em>R</em>)-6′-Methoxycinchonan-9-ol',
'6′-Methoxycinchonidine',
'(-)-Quinine',
'(8<em>S</em>,9<em>R</em>)-Quinine',
'(<em>R</em>)-(-)-Quinine',
'NSC 192949',
'WR297608',
'Qualaquin',
'Mosgard',
'Quinlup',
'Quine 9',
'Cinkona',
'Quinex',
'Quinlex',
'Rezquin',
'QSM',
'SW 85833',
'(<em>R</em>)-(6-Methoxy-4-quinolyl)[(2<em>S</em>)-5-vinylquinuclidin-2-yl]methanol',
'MeSH ID: D011803'],
'uri': 'substance/pt/130950'}
Handle multiple results#
# setup search query parameters
search_base_url = "https://commonchemistry.cas.org/api/search?q="
# SMILES for butadiene
smi_bd = "C=CC=C"
# Request data from CAS Common Chemistry Search API
smi_search_data = requests.get(search_base_url + smi_bd).json()
# get results count
print(smi_search_data["count"])
7
# extract out CAS RNs
smi_casrn_list = []
for casrn_idx in range(len(smi_search_data["results"])):
smi_casrn_list.append(smi_search_data["results"][casrn_idx]["rn"])
print(smi_casrn_list)
['106-99-0', '16422-75-6', '26952-74-9', '29406-96-0', '29989-19-3', '31567-90-5', '9003-17-2']
# now use the detail API to retrieve the full records
detail_base_url = "https://commonchemistry.cas.org/api/detail?"
smi_detail_data = []
for casrn in smi_casrn_list:
smi_detail_data.append(requests.get(detail_base_url + "cas_rn=" + casrn).json())
sleep(1) # add a delay between API calls
# Get some specific data such as name from the detail records
names = []
for name_idx in range(len(smi_detail_data)):
names.append(smi_detail_data[name_idx]["name"])
print(names)
['1,3-Butadiene', 'Butadiene trimer', 'Butadiene dimer', '1,3-Butadiene, homopolymer, isotactic', '1,3-Butadiene-<em>1</em>,<em>1</em>,<em>2</em>,<em>3</em>,<em>4</em>,<em>4</em>-<em>d</em><sub>6</sub>, homopolymer', 'Syndiotactic polybutadiene', 'Polybutadiene']
Handle multiple page results#
The CAS Common Chemistry API returns 50 results per page, and only the first page is returned by default. If the search returns more than 50 results, the offset option can be added to page through and obtain all results:
# setup search query parameters
search_base_url = "https://commonchemistry.cas.org/api/search?q="
n = "selen*"
# get results count for CAS Common Chemistry Search
num_Results = requests.get(search_base_url + n).json()["count"]
print(num_Results)
192
# Request data and save to a list in a loop for each page
n_search_data = []
for page_idx in range(int(num_Results/50 +1)): # creates [0,1,2,3] for 4 pages
page_data = requests.get(search_base_url + n + "&offset=" + str(page_idx*50)).json()
sleep(1)
n_search_data.append(page_data)
# length of search data includes a top level list for each query
len(n_search_data)
4
# lists within lists contain the results
for idx in range(len(n_search_data)):
print(len(n_search_data[idx]["results"]))
50
50
50
42
# We can index and extract out the first casrn like this
pprint(n_search_data[0]["results"][0]["rn"])
'10025-68-0'
# extract out all CAS RNs from the list of lists
n_casrn_list = []
for n_idx in range(len(n_search_data)): # top level list
for casrn_idx in range(len(n_search_data[n_idx]["results"])): # lists within top level
n_casrn_list.append(n_search_data[n_idx]["results"][casrn_idx]["rn"])
len(n_casrn_list)
192
# show first 10
pprint(n_casrn_list[0:10])
['10025-68-0',
'10026-03-6',
'10026-23-0',
'10101-96-9',
'10102-18-8',
'10102-23-5',
'10112-94-4',
'10161-84-9',
'10214-40-1',
'10236-58-5']
# now we can loop through each casrn and use the detail API to obtain the entire record
# this will query CAS Common Chem 192 times and take ~ 5 min.
detail_base_url = "https://commonchemistry.cas.org/api/detail?"
n_detail_data = []
for casrn in n_casrn_list:
n_detail_data.append(requests.get(detail_base_url + "cas_rn=" + casrn).json())
sleep(1) # !! add a delay between API calls
# Extract out some data such as molecularMass
mms = []
for mm_idx in range(len(n_detail_data)):
mms.append(n_detail_data[mm_idx]["molecularMass"])
len(mms)
192
# view first 20
# note that several do not have molecularMass values and have an empty string in the record
print(mms[0:20])
['228.83', '220.77', '', '', '', '', '', '300.24', '', '168.05', '', '', '', '', '', '241.11', '', '368.25', '265.00', '']
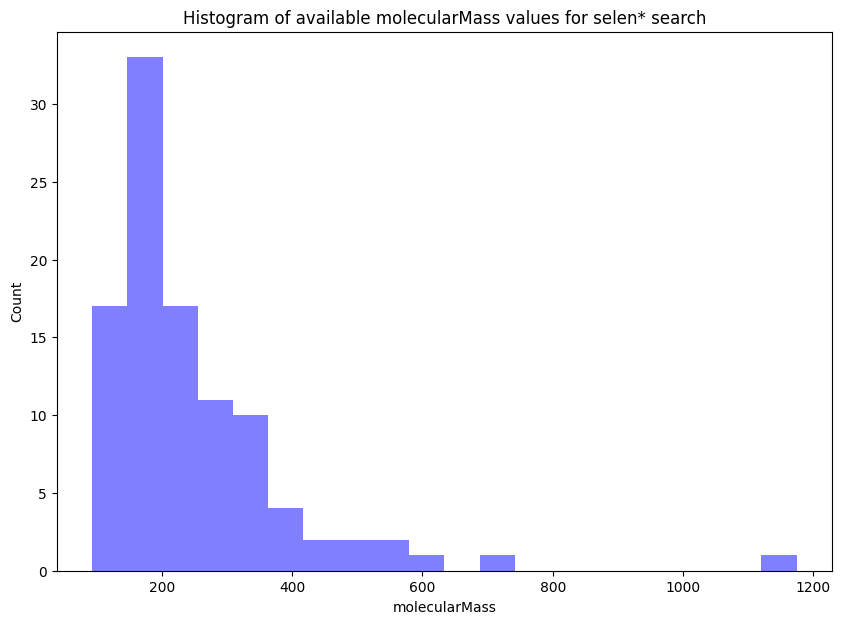
# finally, we can quickly create a simple visualization from the
# extracted molecularMass values (from the selen* search)
# remove empty strings
mms_values = list(filter(None, mms))
# convert to floats
mms_values_float = []
for mms_value in mms_values:
mms_values_float.append(float(mms_value))
# import numpy and matplotlib
import matplotlib.pyplot as plt
# plot data
plt.figure(figsize=(10,7))
plt.hist(mms_values_float, histtype='bar',bins = 20, facecolor="blue", alpha=0.5)
plt.title("Histogram of available molecularMass values for selen* search")
plt.xlabel("molecularMass")
plt.ylabel("Count")
plt.show()