Using the OPSIN API in Python#
About this interactive  recipe
recipe
Author(s): Stuart Chalk
Reviewer(s): Jordi Cuadros
Topic(s): OPSIN API, Chemical identifiers, Chemical images, IUPAC compound names
Format(s): Interactive Jupyter Notebook (Python)
Scenario(s): I need to access chemical identifiers an/or chemical images using code
Skill(s): You should be familiar with
Learning outcomes: After completing this example you should understand:
How to write Python code to request data from a URL (typically an API)
How to use a Python variable to dynamically (with different values) call an API
How to access an image file from the OPSIN image API
How to use regular expressions (regex) to extract data from strings
Citation: ‘Using the OPSIN API in Python’, IUPAC FAIR Chemistry Cookbook, https://w3id.org/ifcc/IFCC002
Reuse: This notebook is made available under a CC-BY-4.0 license.
About OPSIN#
OPSIN is a web tool for converting an IUPAC systematic name into chemical identifiers, chemical markup language (CML) XML and images of molecules. It is written in Java (GitHub repository) and accessed via a web form (for humans), or its API (for machines). More information about OPSIN can be found here (website) or here (paper).
Step 1: Import the Python packages#
The following Python packages are installed in order to run the Python code. Any package imported using ‘from’ is code developed by someone from the Python community (and typically made available to the community via pypi.org), while a package imported starting with ‘import’ is native to Python.
from IPython.display import Image, display # package to run Python in a Jupyter notebook
import requests # package to get data from a URL
import json # package to read/write/display JSON
import re # package to use regular expression (regex) searching
Step 2: Call the OPSIN data API#
Calling an OPSIN API involves adding an IUPAC systematic name to the end of the base OPSIN API endpoint (see the path variable). The format of the API request is ‘https://opsin.ch.cam.ac.uk/opsin/<systematicname>.json’. This call returns data as in JSON format even if the request does not work. If the request does not work the name provided is either no recognized or is not a systematic name. In the example below, the ‘cml’ data in the JSON retrieved is removed to improve the display of the other data.
path = "https://opsin.ch.cam.ac.uk/opsin/" # URL path to the OPSIN API
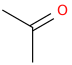
name = "propan-2-one" # IUPAC name of a chemical compound, ion or element
apiurl = path + name + '.json' # concatenate (join) strings with the '+' operator
reqdata = requests.get(apiurl) # get is a method of request data from the OPSIN server
jsondata = reqdata.json() # get the downloaded JSON
del jsondata['cml'] # remove the cml element of the JSON for nicer display
print(apiurl) # print out the apiurl (useful as a check if an error is returned)
print(json.dumps(jsondata, indent=4)) # print the JSON in a nice format
https://opsin.ch.cam.ac.uk/opsin/propan-2-one.json
{
"status": "SUCCESS",
"message": "",
"inchi": "InChI=1/C3H6O/c1-3(2)4/h1-2H3",
"stdinchi": "InChI=1S/C3H6O/c1-3(2)4/h1-2H3",
"stdinchikey": "CSCPPACGZOOCGX-UHFFFAOYSA-N",
"smiles": "CC(C)=O"
}
Step 3: Call the OPSIN image API#
To request an image, rather than the JSON data (above), appending ‘.png’ (portable network graphic) or ‘.svg’ (scaled vector graphics) instead of ‘.json’ will send back and image of a molecule if the name can be interpretted. The format of the API request is ‘https://opsin.ch.cam.ac.uk/opsin/<systematicname>.png’ for a ‘.png’ file and ‘https://opsin.ch.cam.ac.uk/opsin/<systematicname>.svg’ for an ‘.svg’ file.
NOTE: Other options for images can be found at the Chemical Identifier Resolver from the US NIH, see this blog https://cactus.nci.nih.gov/blog/?p=136.
reqimg = requests.get(path + name + ".png") # request the image of the compound
display(Image(reqimg.content)) # display the image
Step 4: Extract the formula of the substance#
An InChI string contains the molecular formula of the compound as part of the string. Using regular expressions (formatted strings that match patterns in other text strings), also referred to as ‘regex’, you can find patterns in strings and extract them, or use them to create new strings. Below, the ‘1S’ part of a standard InChI string is used to anchor a regular expression to match the molecular formula (see note). The string “(.+)” means ‘match any character multiple times, in a sequence until you find a ‘)’ character’. The ‘?’ is required to stop the regex from being ‘greedy’, matching the string all the way to the last ‘/’ in the InChI string, rather than stopping at the first, like we want.
Note
For ionic compounds and salts of organics the InChI code adds their formula in the form ‘cation.anion’, e.g., ethylammonium nitrate InChI=1S/C2H7N.NO3/c1-2-3;2-1(3)4/h2-3H2,1H3;/q;-1/p+1). This is not the normal format for a molecular formula, but you can generate one by totaling each element. Also note that the cation and anion formulae do not include the charges. That is represented in the charge layer (q) at the end of the InChI.
print('InchI: ' + jsondata['stdinchi']) # print the standard inchi
match = re.findall('1S/(.+?)/', jsondata['stdinchi']) # match the formula using regex string
print('Formula: ' + match[0]) # print the first (only) match
InchI: InChI=1S/C3H6O/c1-3(2)4/h1-2H3
Formula: C3H6O
Step 5: Try other queries#
By changing the value of the ‘name’ variable in Step 2 and rerunning Steps 2, 3 and 4, you can retrieve data, get the image and formula for other molecules. Try a molecule that is larger like ‘2-[3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-4-methyl-1,3-thiazol-3-ium-5-yl]ethanol’. If you delete (or comment out line 6 in the code of Step 2 by putting a ‘# ‘ in front of the code) you will also see the CML XML in the output.


