Computing InChIs#
About this interactive  recipe
recipe
Author: Vincent Scalfani
Reviewer: Stuart Chalk
Topics: How to Calculate InChIs from SMILES, Using RDKit or Open Babel Adapted from CPCDS 2021 Digital IUPAC Session - 51st IUPAC General Assembly
Format: Interactive Jupyter Notebook (Python)
Scenarios: You need to convert a SMILES string into its equivalent InChI string.
Skills: You should be familiar with
Learning outcomes: After completing this example you should understand:
How to load and use RDKit to obtain and display chemical identifiers
How to load and use Open Babel to obtain and display chemical identifiers
Citation: ‘Computing InChIs’, Vincent Scalfani, The IUPAC FAIR Chemistry Cookbook, Contributed: 2024-02-14 https://w3id.org/ifcc/IFCC012.
Reuse: This notebook is made available under a CC-BY-4.0 license.
1. Using RDKit#
1.1 Import RDKit Modules#
from rdkit import Chem
from rdkit.Chem import Draw
1.2 Create a Molecular Object from SMILES#
# PubChem CID: 134601
m = Chem.MolFromSmiles('COC(=O)[C@H](CC1=CC=CC=C1)NC(=O)[C@H](CC(=O)O)N')
m # to show image of molecule
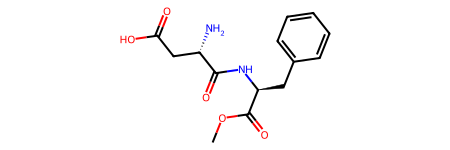
# Internally, we have created an RDKit molecular object
print(m)
<rdkit.Chem.rdchem.Mol object at 0x7ffa2d1fc270>
1.3 Calculate InChI#
# Compute InChI from RDKit mol
Chem.MolToInchi(m)
'InChI=1S/C14H18N2O5/c1-21-14(20)11(7-9-5-3-2-4-6-9)16-13(19)10(15)8-12(17)18/h2-6,10-11H,7-8,15H2,1H3,(H,16,19)(H,17,18)/t10-,11-/m0/s1'
# Compute InChIKey from RDKit mol
Chem.MolToInchiKey(m)
'IAOZJIPTCAWIRG-QWRGUYRKSA-N'
1.4 Calculate InChIs for a List of Molecules#
# Import a file of SMILES strings
smiles_list = []
with open('../files/my_smiles.smi') as infile:
for smi in infile:
smiles_list.append(smi.rstrip()) # rstrip removes newline
print(smiles_list)
['COC(=O)[C@H](CC1=CC=CC=C1)NC(=O)[C@H](CC(=O)O)N', 'COC(=O)[C@@H](CC1=CC=CC=C1)NC(=O)[C@@H](CC(=O)O)N', 'COC(=O)[C@H](CC1=CC=CC=C1)NC(=O)C[C@@H](C(=O)O)N', 'C1=CC=C(C=C1)C[C@@H](C(=O)O)NC(=O)[C@H](CC(=O)O)NC=O', 'C[C@@H](C(=O)N[C@@H](CC1=CC=CC=C1)C(=O)O)N', 'CC(C)C[C@@H](C(=O)N[C@@H](CC1=CC=CC=C1)C(=O)O)NC(=O)C']
# Or create a list directly
smiles_list = ['COC(=O)[C@H](CC1=CC=CC=C1)NC(=O)[C@H](CC(=O)O)N',
'COC(=O)[C@@H](CC1=CC=CC=C1)NC(=O)[C@@H](CC(=O)O)N',
'COC(=O)[C@H](CC1=CC=CC=C1)NC(=O)C[C@@H](C(=O)O)N',
'C1=CC=C(C=C1)C[C@@H](C(=O)O)NC(=O)[C@H](CC(=O)O)NC=O',
'C[C@@H](C(=O)N[C@@H](CC1=CC=CC=C1)C(=O)O)N',
'CC(C)C[C@@H](C(=O)N[C@@H](CC1=CC=CC=C1)C(=O)O)NC(=O)C']
# Next, loop through the smiles_list and create RDKit molecular objects
mols = []
for smi in smiles_list:
mols.append(Chem.MolFromSmiles(smi))
print(mols)
# alternative solution
# mols = [Chem.MolFromSmiles(smi) for smi in smiles_list]
[<rdkit.Chem.rdchem.Mol object at 0x7ffa2d1fc820>, <rdkit.Chem.rdchem.Mol object at 0x7ffa2d1fc900>, <rdkit.Chem.rdchem.Mol object at 0x7ffa2d1fcc80>, <rdkit.Chem.rdchem.Mol object at 0x7ffa2d1fccf0>, <rdkit.Chem.rdchem.Mol object at 0x7ffa2d1fcd60>, <rdkit.Chem.rdchem.Mol object at 0x7ffa2d1fcdd0>]
# Display the molecules in a grid
# SVG False uses PNG
Draw.MolsToGridImage(mols, molsPerRow=3, useSVG=False)
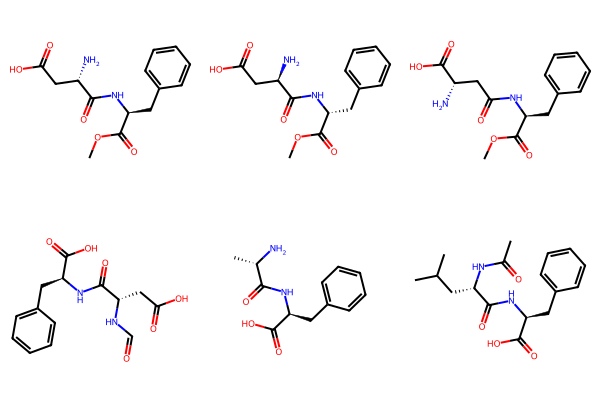
# Loop through mols (molecular objects) and calculate InChIs
InChIs = [Chem.MolToInchi(mol) for mol in mols]
print(InChIs)
['InChI=1S/C14H18N2O5/c1-21-14(20)11(7-9-5-3-2-4-6-9)16-13(19)10(15)8-12(17)18/h2-6,10-11H,7-8,15H2,1H3,(H,16,19)(H,17,18)/t10-,11-/m0/s1', 'InChI=1S/C14H18N2O5/c1-21-14(20)11(7-9-5-3-2-4-6-9)16-13(19)10(15)8-12(17)18/h2-6,10-11H,7-8,15H2,1H3,(H,16,19)(H,17,18)/t10-,11-/m1/s1', 'InChI=1S/C14H18N2O5/c1-21-14(20)11(7-9-5-3-2-4-6-9)16-12(17)8-10(15)13(18)19/h2-6,10-11H,7-8,15H2,1H3,(H,16,17)(H,18,19)/t10-,11-/m0/s1', 'InChI=1S/C14H16N2O6/c17-8-15-10(7-12(18)19)13(20)16-11(14(21)22)6-9-4-2-1-3-5-9/h1-5,8,10-11H,6-7H2,(H,15,17)(H,16,20)(H,18,19)(H,21,22)/t10-,11-/m0/s1', 'InChI=1S/C12H16N2O3/c1-8(13)11(15)14-10(12(16)17)7-9-5-3-2-4-6-9/h2-6,8,10H,7,13H2,1H3,(H,14,15)(H,16,17)/t8-,10-/m0/s1', 'InChI=1S/C17H24N2O4/c1-11(2)9-14(18-12(3)20)16(21)19-15(17(22)23)10-13-7-5-4-6-8-13/h4-8,11,14-15H,9-10H2,1-3H3,(H,18,20)(H,19,21)(H,22,23)/t14-,15-/m0/s1']
2. Using Open Babel#
2.1 Import Open Babel Modules#
# Open Babel v3.1.1
from openbabel import pybel
2.2 Create a Molecular Object from SMILES#
m = pybel.readstring("smi", "COC(=O)[C@H](CC1=CC=CC=C1)NC(=O)[C@H](CC(=O)O)N")
m # to show image of molecule
# Internally, we have created an Open Babel molecular object
print(type(m))
<class 'openbabel.pybel.Molecule'>
2.3 Calculate InChI#
# Set up InChI conversion
conv = pybel.ob.OBConversion()
conv.SetOutFormat("inchi")
True
# Calculate InChI
inchi_output = conv.WriteString(m.OBMol)
print(inchi_output)
InChI=1S/C14H18N2O5/c1-21-14(20)11(7-9-5-3-2-4-6-9)16-13(19)10(15)8-12(17)18/h2-6,10-11H,7-8,15H2,1H3,(H,16,19)(H,17,18)/t10-,11-/m0/s1
# Set up InChIKey conversion
conv = pybel.ob.OBConversion()
conv.SetOutFormat("inchikey")
True
# Calculate InChIKey
inchikey_output = conv.WriteString(m.OBMol)
print(inchikey_output)
IAOZJIPTCAWIRG-QWRGUYRKSA-N
2.4 Calculate InChIs for a List of Molecules#
# Import a file of SMILES
smiles_list =[]
with open('../files/my_smiles.smi') as infile:
for smi in infile:
smiles_list.append(smi.rstrip()) # rstrip removes newline
print(smiles_list)
['COC(=O)[C@H](CC1=CC=CC=C1)NC(=O)[C@H](CC(=O)O)N', 'COC(=O)[C@@H](CC1=CC=CC=C1)NC(=O)[C@@H](CC(=O)O)N', 'COC(=O)[C@H](CC1=CC=CC=C1)NC(=O)C[C@@H](C(=O)O)N', 'C1=CC=C(C=C1)C[C@@H](C(=O)O)NC(=O)[C@H](CC(=O)O)NC=O', 'C[C@@H](C(=O)N[C@@H](CC1=CC=CC=C1)C(=O)O)N', 'CC(C)C[C@@H](C(=O)N[C@@H](CC1=CC=CC=C1)C(=O)O)NC(=O)C']
# Next,loop through the smiles_list and create OB molecular objects
ms = [pybel.readstring("smi", m) for m in smiles_list]
print(ms)
[<openbabel.pybel.Molecule object at 0x7ffa705ad1d0>, <openbabel.pybel.Molecule object at 0x7ffa2b949610>, <openbabel.pybel.Molecule object at 0x7ffa2b9497d0>, <openbabel.pybel.Molecule object at 0x7ffa2b949650>, <openbabel.pybel.Molecule object at 0x7ffa2b949890>, <openbabel.pybel.Molecule object at 0x7ffa2b949a50>]
# Set up InChI conversion
conv = pybel.ob.OBConversion()
conv.SetOutFormat("inchi")
# Loop through mols (molecular objects) and calculate InChIs
InChIs = [conv.WriteString(m.OBMol).rstrip() for m in ms]
print(InChIs)
['InChI=1S/C14H18N2O5/c1-21-14(20)11(7-9-5-3-2-4-6-9)16-13(19)10(15)8-12(17)18/h2-6,10-11H,7-8,15H2,1H3,(H,16,19)(H,17,18)/t10-,11-/m0/s1', 'InChI=1S/C14H18N2O5/c1-21-14(20)11(7-9-5-3-2-4-6-9)16-13(19)10(15)8-12(17)18/h2-6,10-11H,7-8,15H2,1H3,(H,16,19)(H,17,18)/t10-,11-/m1/s1', 'InChI=1S/C14H18N2O5/c1-21-14(20)11(7-9-5-3-2-4-6-9)16-12(17)8-10(15)13(18)19/h2-6,10-11H,7-8,15H2,1H3,(H,16,17)(H,18,19)/t10-,11-/m0/s1', 'InChI=1S/C14H16N2O6/c17-8-15-10(7-12(18)19)13(20)16-11(14(21)22)6-9-4-2-1-3-5-9/h1-5,8,10-11H,6-7H2,(H,15,17)(H,16,20)(H,18,19)(H,21,22)/t10-,11-/m0/s1', 'InChI=1S/C12H16N2O3/c1-8(13)11(15)14-10(12(16)17)7-9-5-3-2-4-6-9/h2-6,8,10H,7,13H2,1H3,(H,14,15)(H,16,17)/t8-,10-/m0/s1', 'InChI=1S/C17H24N2O4/c1-11(2)9-14(18-12(3)20)16(21)19-15(17(22)23)10-13-7-5-4-6-8-13/h4-8,11,14-15H,9-10H2,1-3H3,(H,18,20)(H,19,21)(H,22,23)/t14-,15-/m0/s1']
References
[1] RDKit Documentation: https://www.rdkit.org/docs/index.html
[2] Open Babel Python Documentation: https://open-babel.readthedocs.io/en/latest/UseTheLibrary/Python.html


